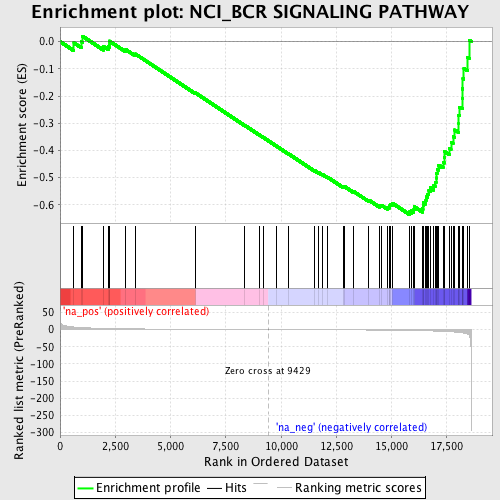
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | NCI_BCR SIGNALING PATHWAY |
| Enrichment Score (ES) | -0.6353433 |
| Normalized Enrichment Score (NES) | -1.8222914 |
| Nominal p-value | 0.009983361 |
| FDR q-value | 0.4940343 |
| FWER p-Value | 0.992 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CAMK2G | 626 | 7.427 | -0.0049 | No | ||
| 2 | MAP3K7 | 972 | 5.905 | -0.0006 | No | ||
| 3 | MALT1 | 1006 | 5.798 | 0.0201 | No | ||
| 4 | IKBKB | 1959 | 3.365 | -0.0181 | No | ||
| 5 | MAPK8 | 2179 | 3.027 | -0.0182 | No | ||
| 6 | CHUK | 2221 | 2.966 | -0.0089 | No | ||
| 7 | CSNK2A1 | 2228 | 2.950 | 0.0022 | No | ||
| 8 | AKT1 | 2974 | 2.198 | -0.0294 | No | ||
| 9 | MAP2K1 | 3408 | 1.872 | -0.0455 | No | ||
| 10 | CALM1 | 6121 | 0.834 | -0.1884 | No | ||
| 11 | POU2F2 | 8349 | 0.267 | -0.3074 | No | ||
| 12 | FOS | 9018 | 0.107 | -0.3430 | No | ||
| 13 | HRAS | 9216 | 0.053 | -0.3534 | No | ||
| 14 | LYN | 9815 | -0.105 | -0.3852 | No | ||
| 15 | SOS1 | 10324 | -0.226 | -0.4117 | No | ||
| 16 | NFKBIB | 11524 | -0.516 | -0.4744 | No | ||
| 17 | SH3BP5 | 11703 | -0.563 | -0.4818 | No | ||
| 18 | PIK3R1 | 11865 | -0.608 | -0.4881 | No | ||
| 19 | IBTK | 12104 | -0.677 | -0.4983 | No | ||
| 20 | RASA1 | 12828 | -0.895 | -0.5338 | No | ||
| 21 | CALM2 | 12857 | -0.904 | -0.5318 | No | ||
| 22 | PAG1 | 13288 | -1.046 | -0.5509 | No | ||
| 23 | PPP3CA | 13975 | -1.312 | -0.5828 | No | ||
| 24 | BTK | 14460 | -1.507 | -0.6030 | No | ||
| 25 | MAPK14 | 14537 | -1.541 | -0.6012 | No | ||
| 26 | RAF1 | 14806 | -1.685 | -0.6091 | No | ||
| 27 | SHC1 | 14889 | -1.736 | -0.6067 | No | ||
| 28 | RAC1 | 14926 | -1.756 | -0.6019 | No | ||
| 29 | CD19 | 14965 | -1.777 | -0.5970 | No | ||
| 30 | VAV2 | 15039 | -1.820 | -0.5939 | No | ||
| 31 | CD22 | 15809 | -2.447 | -0.6258 | Yes | ||
| 32 | CARD11 | 15904 | -2.542 | -0.6210 | Yes | ||
| 33 | ETS1 | 16011 | -2.677 | -0.6164 | Yes | ||
| 34 | MAP4K1 | 16033 | -2.697 | -0.6070 | Yes | ||
| 35 | FCGR2B | 16396 | -3.211 | -0.6141 | Yes | ||
| 36 | PIK3CA | 16426 | -3.246 | -0.6031 | Yes | ||
| 37 | PTPRC | 16439 | -3.278 | -0.5910 | Yes | ||
| 38 | IKBKG | 16553 | -3.477 | -0.5836 | Yes | ||
| 39 | CALM3 | 16568 | -3.499 | -0.5708 | Yes | ||
| 40 | GRB2 | 16640 | -3.636 | -0.5605 | Yes | ||
| 41 | CD79A | 16691 | -3.727 | -0.5487 | Yes | ||
| 42 | PPP3CB | 16752 | -3.824 | -0.5371 | Yes | ||
| 43 | PLCG2 | 16913 | -4.165 | -0.5296 | Yes | ||
| 44 | PPP3CC | 16975 | -4.313 | -0.5161 | Yes | ||
| 45 | BLNK | 17018 | -4.436 | -0.5012 | Yes | ||
| 46 | MAP3K1 | 17023 | -4.458 | -0.4841 | Yes | ||
| 47 | RELA | 17065 | -4.586 | -0.4685 | Yes | ||
| 48 | JUN | 17135 | -4.762 | -0.4537 | Yes | ||
| 49 | MAPK3 | 17335 | -5.344 | -0.4437 | Yes | ||
| 50 | NFKB1 | 17383 | -5.487 | -0.4250 | Yes | ||
| 51 | PTEN | 17406 | -5.544 | -0.4046 | Yes | ||
| 52 | NFKBIA | 17625 | -6.306 | -0.3919 | Yes | ||
| 53 | DAPP1 | 17695 | -6.511 | -0.3704 | Yes | ||
| 54 | MAPK1 | 17787 | -6.868 | -0.3486 | Yes | ||
| 55 | DOK1 | 17867 | -7.248 | -0.3248 | Yes | ||
| 56 | PDPK1 | 18025 | -8.051 | -0.3020 | Yes | ||
| 57 | BCL10 | 18031 | -8.105 | -0.2708 | Yes | ||
| 58 | SYK | 18076 | -8.451 | -0.2404 | Yes | ||
| 59 | CSK | 18203 | -9.505 | -0.2103 | Yes | ||
| 60 | CD79B | 18213 | -9.597 | -0.1735 | Yes | ||
| 61 | TRAF6 | 18232 | -9.851 | -0.1363 | Yes | ||
| 62 | NFATC1 | 18277 | -10.197 | -0.0991 | Yes | ||
| 63 | PTPN6 | 18441 | -13.086 | -0.0571 | Yes | ||
| 64 | CD72 | 18518 | -17.128 | 0.0053 | Yes |